Exploratory Data Analysis Basics#
EDA Definition#

Razones para hacer un EDA#
Organizar y entender las variables
Establecer relaciones entre las variables
Encontrar patrones ocultos en los datos
Ayuda a escoger el modelo correcto para la necesidad correcta
Tomar decisiones informadas
Pasos de EDA#

Tipos de Analitica de datos#

Project#
Environment Setup#
Library installation#
!pip install --upgrade pip
!pip install palmerpenguins==0.1.4 numpy==1.23.4 pandas==1.5.1 seaborn==0.12.1 matplotlib==3.6.0 empiricaldist==0.6.7 statsmodels==0.13.5 scikit-learn==1.1.2 pyjanitor==0.23.1 session-info
Requirement already satisfied: pip in c:\users\admin\miniconda3\envs\jupyter_notebooks_from_deepnote\lib\site-packages (23.2.1)
^C
Collecting palmerpenguins==0.1.4
Using cached palmerpenguins-0.1.4-py3-none-any.whl (17 kB)
Collecting numpy==1.23.4
Using cached numpy-1.23.4-cp311-cp311-win_amd64.whl (14.6 MB)
Collecting pandas==1.5.1
Using cached pandas-1.5.1-cp311-cp311-win_amd64.whl (10.3 MB)
Collecting seaborn==0.12.1
Using cached seaborn-0.12.1-py3-none-any.whl (288 kB)
Collecting matplotlib==3.6.0
Using cached matplotlib-3.6.0-cp311-cp311-win_amd64.whl (7.2 MB)
Collecting empiricaldist==0.6.7
Using cached empiricaldist-0.6.7.tar.gz (11 kB)
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Collecting statsmodels==0.13.5
Using cached statsmodels-0.13.5-cp311-cp311-win_amd64.whl (9.0 MB)
Collecting scikit-learn==1.1.2
Using cached scikit-learn-1.1.2.tar.gz (7.0 MB)
Installing build dependencies: started
Library import#
import empiricaldist
import janitor
import matplotlib.pyplot as plt
import numpy as np
import palmerpenguins
import pandas as pd
import scipy.stats
import seaborn as sns
import sklearn.metrics
import statsmodels.api as sm
import statsmodels.formula.api as smf
import statsmodels.stats as ss
import session_info
Stablish Chart Settings#
%matplotlib inline
sns.set_style(style='whitegrid')
sns.set_context(context='notebook')
plt.rcParams['figure.figsize'] = (11, 9.4)
penguin_color = {
'Adelie': '#ff6602ff',
'Gentoo': '#0f7175ff',
'Chinstrap': '#c65dc9ff'
}
Importing Dataset#
The dataset chosen for this project is the palmerpenguins dataset
The goal of the Palmer Penguins dataset is to replace the highly overused Iris dataset for data exploration & visualization.
The information contains size measurements, clutch observations, and blood isotope ratios for 344 adult foraging Adélie, Chinstrap, and Gentoo penguins observed on islands in the Palmer Archipelago near Palmer Station, Antarctica.
Data were collected and made available by Dr. Kristen Gorman and the Palmer Station, Antarctica Long Term Ecological Research (LTER) Program.
Raw Data#
This package contains a raw dataset (literally contains the data as it was collected)
raw_df = palmerpenguins.load_penguins_raw()
raw_df
| studyName | Sample Number | Species | Region | Island | Stage | Individual ID | Clutch Completion | Date Egg | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | Comments | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | PAL0708 | 1 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A1 | Yes | 2007-11-11 | 39.1 | 18.7 | 181.0 | 3750.0 | MALE | NaN | NaN | Not enough blood for isotopes. |
| 1 | PAL0708 | 2 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A2 | Yes | 2007-11-11 | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 | NaN |
| 2 | PAL0708 | 3 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A1 | Yes | 2007-11-16 | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 | NaN |
| 3 | PAL0708 | 4 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A2 | Yes | 2007-11-16 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | Adult not sampled. |
| 4 | PAL0708 | 5 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N3A1 | Yes | 2007-11-16 | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | PAL0910 | 64 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N98A2 | Yes | 2009-11-19 | 55.8 | 19.8 | 207.0 | 4000.0 | MALE | 9.70465 | -24.53494 | NaN |
| 340 | PAL0910 | 65 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N99A1 | No | 2009-11-21 | 43.5 | 18.1 | 202.0 | 3400.0 | FEMALE | 9.37608 | -24.40753 | Nest never observed with full clutch. |
| 341 | PAL0910 | 66 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N99A2 | No | 2009-11-21 | 49.6 | 18.2 | 193.0 | 3775.0 | MALE | 9.46180 | -24.70615 | Nest never observed with full clutch. |
| 342 | PAL0910 | 67 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N100A1 | Yes | 2009-11-21 | 50.8 | 19.0 | 210.0 | 4100.0 | MALE | 9.98044 | -24.68741 | NaN |
| 343 | PAL0910 | 68 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N100A2 | Yes | 2009-11-21 | 50.2 | 18.7 | 198.0 | 3775.0 | FEMALE | 9.39305 | -24.25255 | NaN |
344 rows × 17 columns
processed data#
Dataset containing already processed data
df = palmerpenguins.load_penguins()
df
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
344 rows × 8 columns
Using the seaborn dataset#
Seaborn also contains this dataset, let’s see it:
sns.load_dataset("penguins")
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Gentoo | Biscoe | NaN | NaN | NaN | NaN | NaN |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
344 rows × 7 columns
Performing EDA#
Now let’s answer some questions, remember that the name of our dataframe is stored in df
What are the variables types for this dataset?#
You can use pandas attribute dtype to do this:
df.dtypes
species object
island object
bill_length_mm float64
bill_depth_mm float64
flipper_length_mm float64
body_mass_g float64
sex object
year int64
dtype: object
“object” is usually synonymous of categorical variables
“float64” is usually synonymous of Numerical continuous variables
“int64” is usually synonymous of Numerical discrete variables
How many variables of each type is present in the dataset?#
The value_counts() attribute can help
(
df
.dtypes
.value_counts()
)
float64 4
object 3
int64 1
dtype: int64
How many variables and records are in the dataset?#
The .shape() function can help with this
df.shape
(344, 8)
344 rows
8 columns
That answers the question
Are there null values in the dataset?#
.isnull() returns boolean values
any() tells you if null values are present in a column
See the result below:
df.isnull().any()
species False
island False
bill_length_mm True
bill_depth_mm True
flipper_length_mm True
body_mass_g True
sex True
year False
dtype: bool
How many null values are there in each column?#
use .sum() to obtain the results
df.isnull().sum()
species 0
island 0
bill_length_mm 2
bill_depth_mm 2
flipper_length_mm 2
body_mass_g 2
sex 11
year 0
dtype: int64
What is the total amount of null values in the dataset?#
call .sum() again in the previous result
df.isnull().sum().sum()
19
What is the null values proportion for each variable?#
let’s show it in percentage
( df.isnull().sum()/(df.shape[0]) )*100
species 0.000000
island 0.000000
bill_length_mm 0.581395
bill_depth_mm 0.581395
flipper_length_mm 0.581395
body_mass_g 0.581395
sex 3.197674
year 0.000000
dtype: float64
You can actually make a chart to represent this:
melted = df.isnull().melt()
melted
| variable | value | |
|---|---|---|
| 0 | species | False |
| 1 | species | False |
| 2 | species | False |
| 3 | species | False |
| 4 | species | False |
| ... | ... | ... |
| 2747 | year | False |
| 2748 | year | False |
| 2749 | year | False |
| 2750 | year | False |
| 2751 | year | False |
2752 rows × 2 columns
melted.pipe(
lambda x : sns.displot(data=x, y="variable", hue="value", multiple="fill", aspect=2)
)
<seaborn.axisgrid.FacetGrid at 0x7fa235ed23a0>
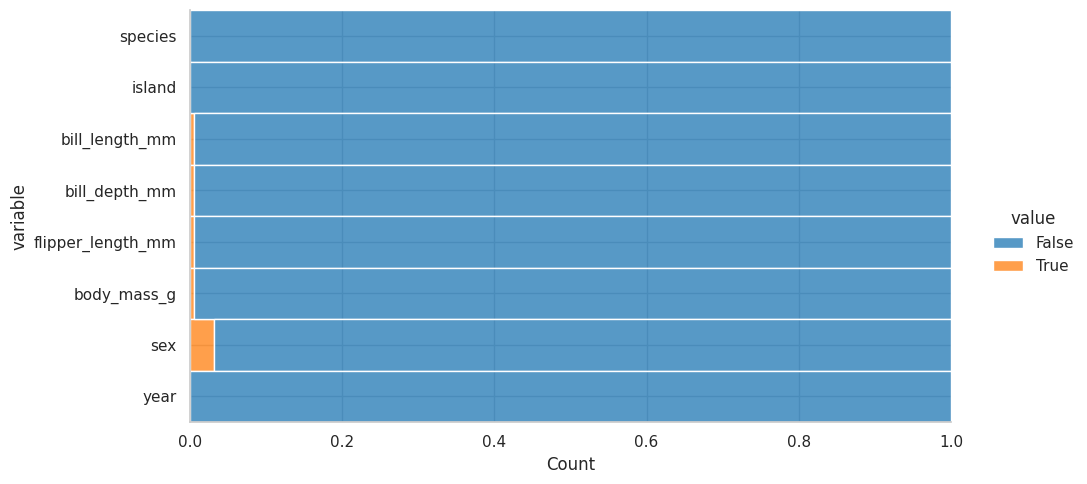
Los valores nulos no llegan ni al 10%
What are the records that contain the null values in the dataset? (Visualize)#
First create the dataframe
nulls = df.isnull().transpose()
nulls
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 334 | 335 | 336 | 337 | 338 | 339 | 340 | 341 | 342 | 343 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| island | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| bill_length_mm | False | False | False | True | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| bill_depth_mm | False | False | False | True | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| flipper_length_mm | False | False | False | True | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| body_mass_g | False | False | False | True | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| sex | False | False | False | True | False | False | False | False | True | True | ... | False | False | False | False | False | False | False | False | False | False |
| year | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
8 rows × 344 columns
Now the chart
plt.figure(figsize=(7, 7))
nulls.pipe(
lambda x : sns.heatmap(data=x)
)
# Alternatively, use:
#df.isnull().transpose().pipe(lambda x : sns.heatmap(data=x))
<AxesSubplot: >
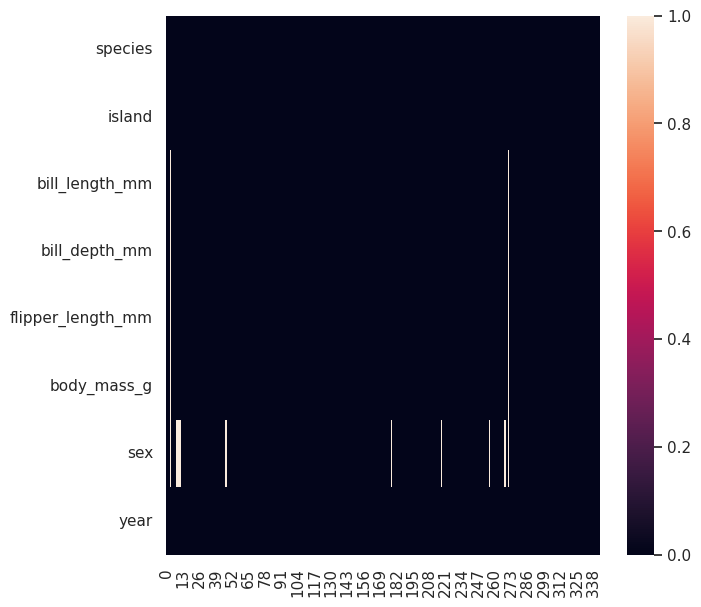
Here you can evidence:
The missing values (Not corresponding to sex) are associated with only two records
The sex missing values are consistant, we should not worry about that
You might want to delete those two records that contain the vast majority of missing values (Not corresponding to sex)
How many records will be lost if we delete all the missing values?#
df.dropna()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | male | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
333 rows × 8 columns
Note that now you have 333 rows, instead of the original 344
THIS COMMAND DOES NOT MODIFY YOUR ORIGINAL DATABASE!!!!
df.shape
(344, 8)
DO YOU SEE?
SO, LET’S SAVE IT into another variable ;)
df_clean = df.dropna()
df_clean
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | male | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
333 rows × 8 columns
Counts and Proportions#
What measures describe our dataset?#
The function .describe() can do this for us, but it will just do it for numerical variables.
To include strings use .describe(include=”all”)
df.describe(include="all")
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| count | 344 | 344 | 342.000000 | 342.000000 | 342.000000 | 342.000000 | 333 | 344.000000 |
| unique | 3 | 3 | NaN | NaN | NaN | NaN | 2 | NaN |
| top | Adelie | Biscoe | NaN | NaN | NaN | NaN | male | NaN |
| freq | 152 | 168 | NaN | NaN | NaN | NaN | 168 | NaN |
| mean | NaN | NaN | 43.921930 | 17.151170 | 200.915205 | 4201.754386 | NaN | 2008.029070 |
| std | NaN | NaN | 5.459584 | 1.974793 | 14.061714 | 801.954536 | NaN | 0.818356 |
| min | NaN | NaN | 32.100000 | 13.100000 | 172.000000 | 2700.000000 | NaN | 2007.000000 |
| 25% | NaN | NaN | 39.225000 | 15.600000 | 190.000000 | 3550.000000 | NaN | 2007.000000 |
| 50% | NaN | NaN | 44.450000 | 17.300000 | 197.000000 | 4050.000000 | NaN | 2008.000000 |
| 75% | NaN | NaN | 48.500000 | 18.700000 | 213.000000 | 4750.000000 | NaN | 2009.000000 |
| max | NaN | NaN | 59.600000 | 21.500000 | 231.000000 | 6300.000000 | NaN | 2009.000000 |
What if i want only numerical variables?
df.describe(include=[np.number])
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | year | |
|---|---|---|---|---|---|
| count | 342.000000 | 342.000000 | 342.000000 | 342.000000 | 344.000000 |
| mean | 43.921930 | 17.151170 | 200.915205 | 4201.754386 | 2008.029070 |
| std | 5.459584 | 1.974793 | 14.061714 | 801.954536 | 0.818356 |
| min | 32.100000 | 13.100000 | 172.000000 | 2700.000000 | 2007.000000 |
| 25% | 39.225000 | 15.600000 | 190.000000 | 3550.000000 | 2007.000000 |
| 50% | 44.450000 | 17.300000 | 197.000000 | 4050.000000 | 2008.000000 |
| 75% | 48.500000 | 18.700000 | 213.000000 | 4750.000000 | 2009.000000 |
| max | 59.600000 | 21.500000 | 231.000000 | 6300.000000 | 2009.000000 |
What if i want only categorical variables?
df.describe(include=object)
| species | island | sex | |
|---|---|---|---|
| count | 344 | 344 | 333 |
| unique | 3 | 3 | 2 |
| top | Adelie | Biscoe | male |
| freq | 152 | 168 | 168 |
BONUS: Transform an object into a category#
See how to transform a column with the data type “object into a category”
This allows cool things such as see the proportions
df.astype({
"species":"category", #just insert a dict where key is column name and value is the type
"island":"category",
"sex":"category"
})
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | male | 2009 |
| 340 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | female | 2009 |
| 341 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | male | 2009 |
| 342 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | male | 2009 |
| 343 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | female | 2009 |
344 rows × 8 columns
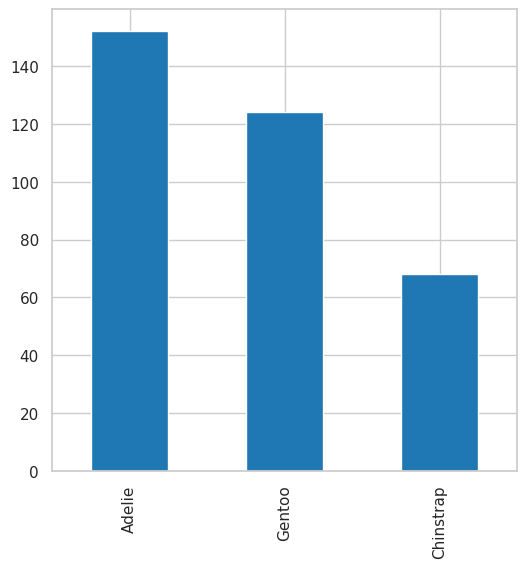
How to visualize counts?#
Pandas
plt.figure(figsize=(6, 6))
df.species.value_counts().plot(kind="bar");
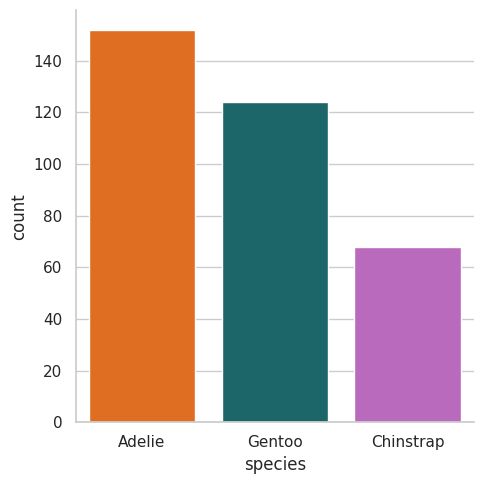
Seaborn
sns.catplot(data=df, x="species", kind="count", palette=penguin_color);
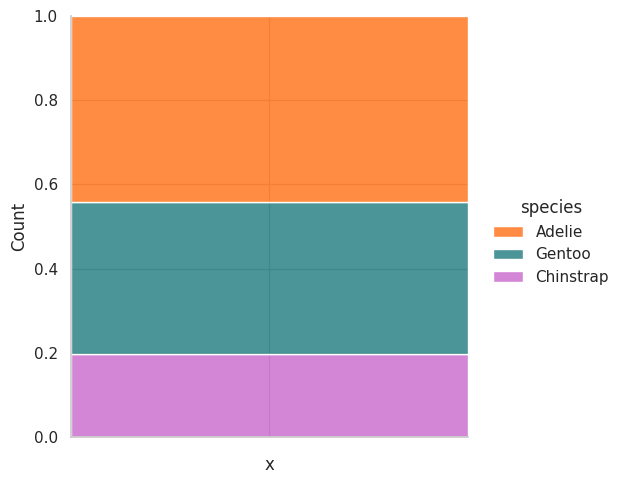
How to visualize proportions#
Seaborn
df.add_column("x", "").pipe(
lambda df: (
sns.displot(data=df, x="x", hue="species", multiple="fill", palette=penguin_color)
)
);
Measures of central tendency#
Mean#
for all the variables
df.mean()
/tmp/ipykernel_1302/3698961737.py:1: FutureWarning: The default value of numeric_only in DataFrame.mean is deprecated. In a future version, it will default to False. In addition, specifying 'numeric_only=None' is deprecated. Select only valid columns or specify the value of numeric_only to silence this warning.
df.mean()
bill_length_mm 43.921930
bill_depth_mm 17.151170
flipper_length_mm 200.915205
body_mass_g 4201.754386
year 2008.029070
dtype: float64
A penguin is around 4 kg LOLLLLLL
Only one variable
df.bill_depth_mm.mean()
17.151169590643274
Median#
df.median()
/tmp/ipykernel_1302/530051474.py:1: FutureWarning: The default value of numeric_only in DataFrame.median is deprecated. In a future version, it will default to False. In addition, specifying 'numeric_only=None' is deprecated. Select only valid columns or specify the value of numeric_only to silence this warning.
df.median()
bill_length_mm 44.45
bill_depth_mm 17.30
flipper_length_mm 197.00
body_mass_g 4050.00
year 2008.00
dtype: float64
Mode#
df.mode()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Biscoe | 41.1 | 17.0 | 190.0 | 3800.0 | male | 2009 |
Works for categorical too
Measures of dispersion#
Maximum value for each variable?#
df.max(numeric_only=True) #numeric_only=True excludes the categorical variables(makes sense)
bill_length_mm 59.6
bill_depth_mm 21.5
flipper_length_mm 231.0
body_mass_g 6300.0
year 2009.0
dtype: float64
Minimum value for each variable?#
df.min(numeric_only=True)
bill_length_mm 32.1
bill_depth_mm 13.1
flipper_length_mm 172.0
body_mass_g 2700.0
year 2007.0
dtype: float64
Range of each variable#
df.max(numeric_only=True) - df.min(numeric_only=True)
bill_length_mm 27.5
bill_depth_mm 8.4
flipper_length_mm 59.0
body_mass_g 3600.0
year 2.0
dtype: float64
Standart deviation#
df.std()
/tmp/ipykernel_1302/3390915376.py:1: FutureWarning: The default value of numeric_only in DataFrame.std is deprecated. In a future version, it will default to False. In addition, specifying 'numeric_only=None' is deprecated. Select only valid columns or specify the value of numeric_only to silence this warning.
df.std()
bill_length_mm 5.459584
bill_depth_mm 1.974793
flipper_length_mm 14.061714
body_mass_g 801.954536
year 0.818356
dtype: float64
Interquartile range?#
The IQR equals Q3-Q1, and it holds the 50% of the data around the mean
Q1
df.quantile(0.25)
/tmp/ipykernel_1302/3656653379.py:1: FutureWarning: The default value of numeric_only in DataFrame.quantile is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
df.quantile(0.25)
bill_length_mm 39.225
bill_depth_mm 15.600
flipper_length_mm 190.000
body_mass_g 3550.000
year 2007.000
Name: 0.25, dtype: float64
Q3
df.quantile(0.75)
/tmp/ipykernel_1302/3799946287.py:1: FutureWarning: The default value of numeric_only in DataFrame.quantile is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
df.quantile(0.75)
bill_length_mm 48.5
bill_depth_mm 18.7
flipper_length_mm 213.0
body_mass_g 4750.0
year 2009.0
Name: 0.75, dtype: float64
IQR
(
df
.quantile(q=[0.75, 0.50, 0.25])
.transpose()
.rename_axis("variable")
.reset_index()
.assign(
iqr=lambda df: df[0.75] - df[0.25]
)
)
/tmp/ipykernel_1302/4097684495.py:2: FutureWarning: The default value of numeric_only in DataFrame.quantile is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
df
| variable | 0.75 | 0.5 | 0.25 | iqr | |
|---|---|---|---|---|---|
| 0 | bill_length_mm | 48.5 | 44.45 | 39.225 | 9.275 |
| 1 | bill_depth_mm | 18.7 | 17.30 | 15.600 | 3.100 |
| 2 | flipper_length_mm | 213.0 | 197.00 | 190.000 | 23.000 |
| 3 | body_mass_g | 4750.0 | 4050.00 | 3550.000 | 1200.000 |
| 4 | year | 2009.0 | 2008.00 | 2007.000 | 2.000 |
Probabilities#
Probability Mass function#
Probability for each value
empiricaldist.Pmf.from_seq(df.flipper_length_mm)
| probs | |
|---|---|
| 172.0 | 0.002924 |
| 174.0 | 0.002924 |
| 176.0 | 0.002924 |
| 178.0 | 0.011696 |
| 179.0 | 0.002924 |
| 180.0 | 0.014620 |
| 181.0 | 0.020468 |
| 182.0 | 0.008772 |
| 183.0 | 0.005848 |
| 184.0 | 0.020468 |
| 185.0 | 0.026316 |
| 186.0 | 0.020468 |
| 187.0 | 0.046784 |
| 188.0 | 0.017544 |
| 189.0 | 0.020468 |
| 190.0 | 0.064327 |
| 191.0 | 0.038012 |
| 192.0 | 0.020468 |
| 193.0 | 0.043860 |
| 194.0 | 0.014620 |
| 195.0 | 0.049708 |
| 196.0 | 0.029240 |
| 197.0 | 0.029240 |
| 198.0 | 0.023392 |
| 199.0 | 0.017544 |
| 200.0 | 0.011696 |
| 201.0 | 0.017544 |
| 202.0 | 0.011696 |
| 203.0 | 0.014620 |
| 205.0 | 0.008772 |
| 206.0 | 0.002924 |
| 207.0 | 0.005848 |
| 208.0 | 0.023392 |
| 209.0 | 0.014620 |
| 210.0 | 0.040936 |
| 211.0 | 0.005848 |
| 212.0 | 0.020468 |
| 213.0 | 0.017544 |
| 214.0 | 0.017544 |
| 215.0 | 0.035088 |
| 216.0 | 0.023392 |
| 217.0 | 0.017544 |
| 218.0 | 0.014620 |
| 219.0 | 0.014620 |
| 220.0 | 0.023392 |
| 221.0 | 0.014620 |
| 222.0 | 0.017544 |
| 223.0 | 0.005848 |
| 224.0 | 0.008772 |
| 225.0 | 0.011696 |
| 226.0 | 0.002924 |
| 228.0 | 0.011696 |
| 229.0 | 0.005848 |
| 230.0 | 0.020468 |
| 231.0 | 0.002924 |
Then, if i want to know that is the specific probability of getting a value:
empiricaldist.Pmf.from_seq(df.flipper_length_mm)(190)
0.06432748538011696
Empirical Cumulative Distribution Function#
This one returns the cumulative probability based on the dataset
plt.figure(figsize=(6, 6))
sns.ecdfplot(data=df, x="flipper_length_mm");
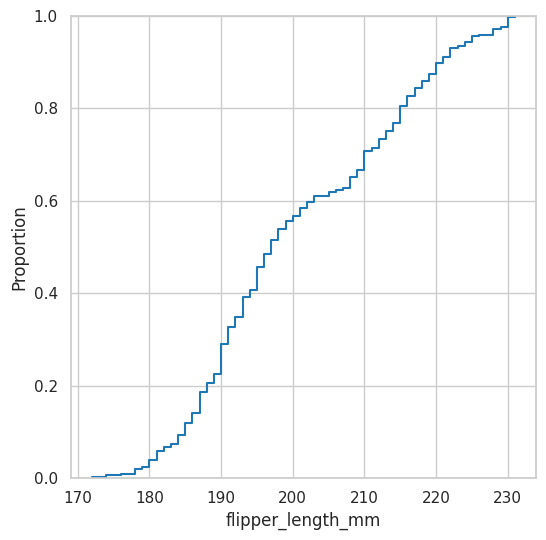
Probabilities of getting a penguin with 200 or less flipper_length?
empiricaldist.Cdf.from_seq(df.flipper_length_mm)(200)
array(0.56725146)
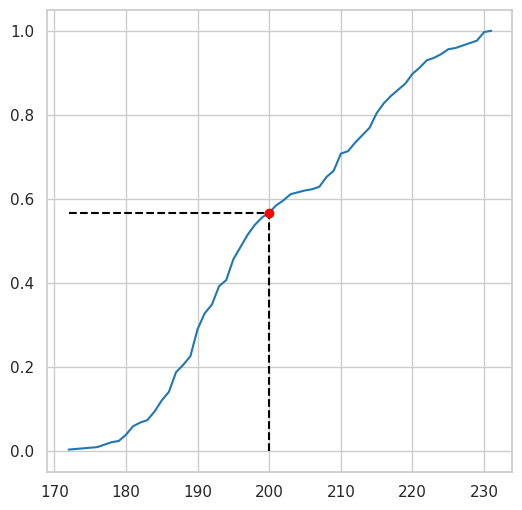
See the chart below for a better interpretation
plt.figure(figsize=(6, 6))
empiricaldist.Cdf.from_seq(df.flipper_length_mm).plot()
q=200
p = empiricaldist.Cdf.from_seq(df.flipper_length_mm)(200)
plt.vlines(
x=q,
ymin=0,
ymax=p,
color="black",
linestyle="dashed"
)
plt.hlines(
y=p,
xmin=empiricaldist.Cdf.from_seq(df.flipper_length_mm).qs[0],
xmax=q,
color="black",
linestyle="dashed"
)
plt.plot(q, p, "ro");
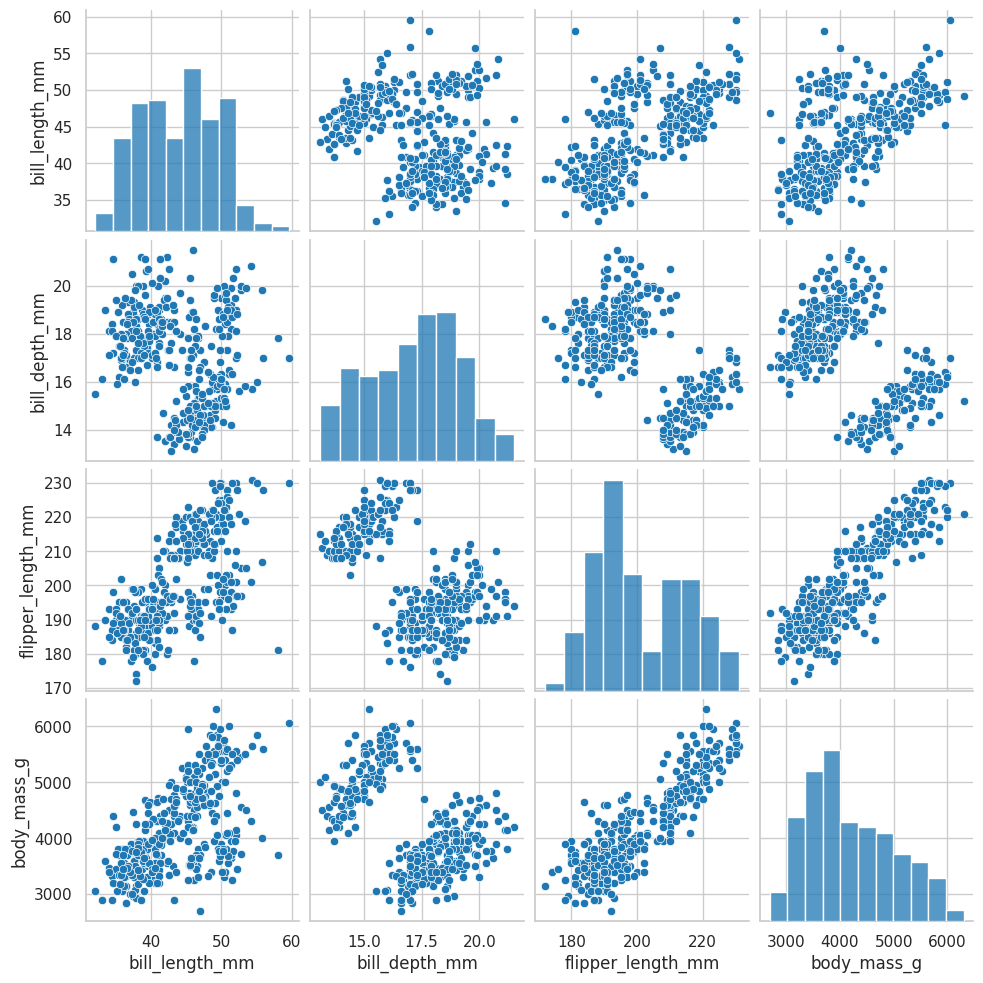
Correlations#
Pairplot#
In this case, please remove the year column, despite being a numeric column, it is not adding value to the correlation analysis
sns.pairplot(data=df.drop(["year"], axis=1));
Pairplot per specie#
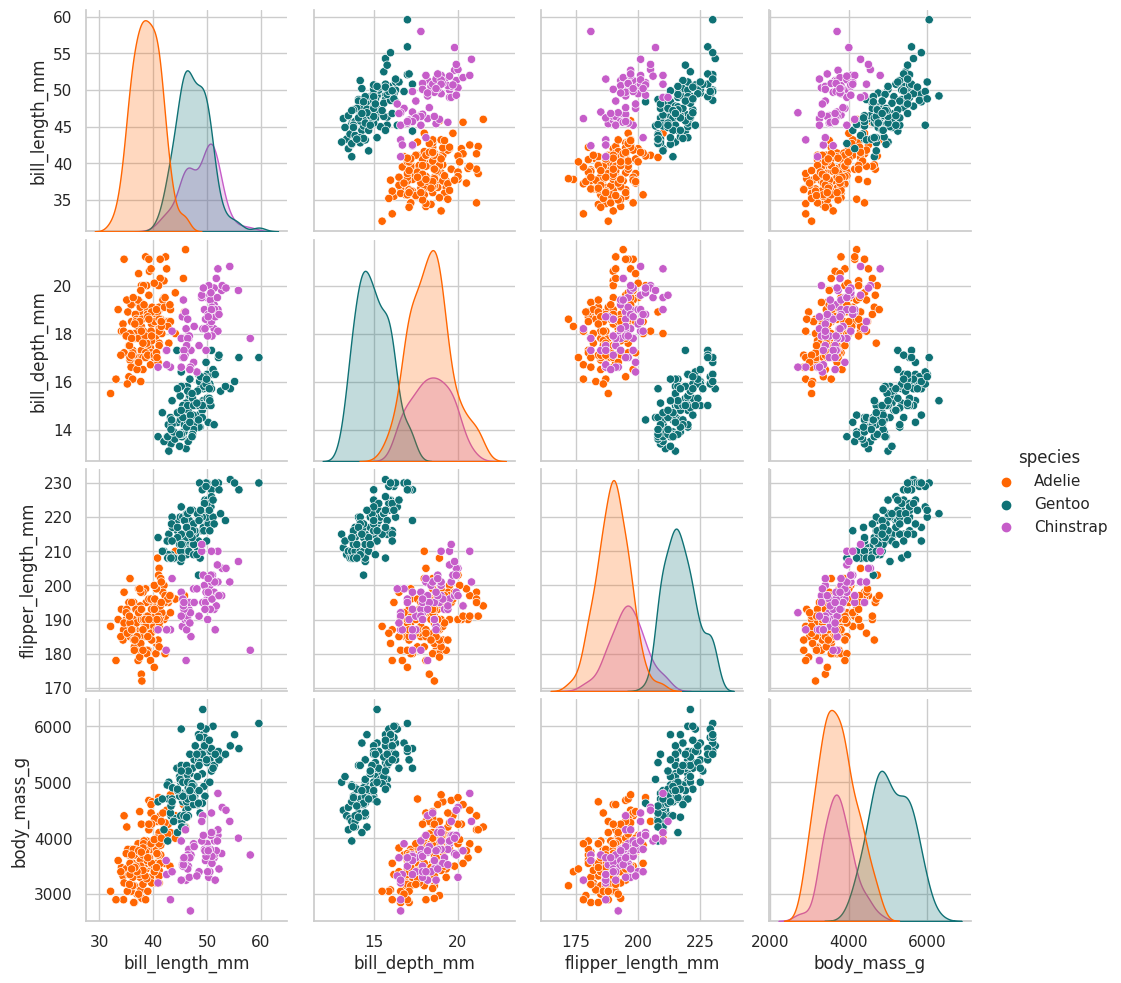
sns.pairplot(data=df.drop(["year"], axis=1), hue="species", palette=penguin_color);
Swarmplot#
This one can be used to associate a categorical variable with a numerical:
plt.figure(figsize=(6, 6))
sns.swarmplot(
data=df,
x="island",
y="body_mass_g",
hue="species",
palette=penguin_color
);
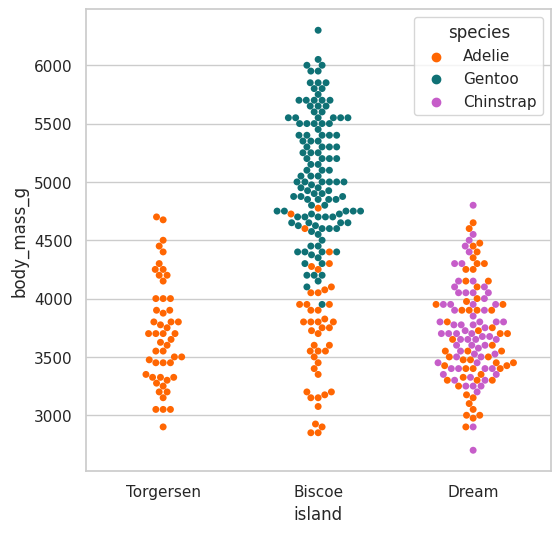
There doesn’t seem to be a relation among the penguin body mass and the island where it is located
there is a relation among the body mass and the specie
Is there a linear correlation among any of our variables?#
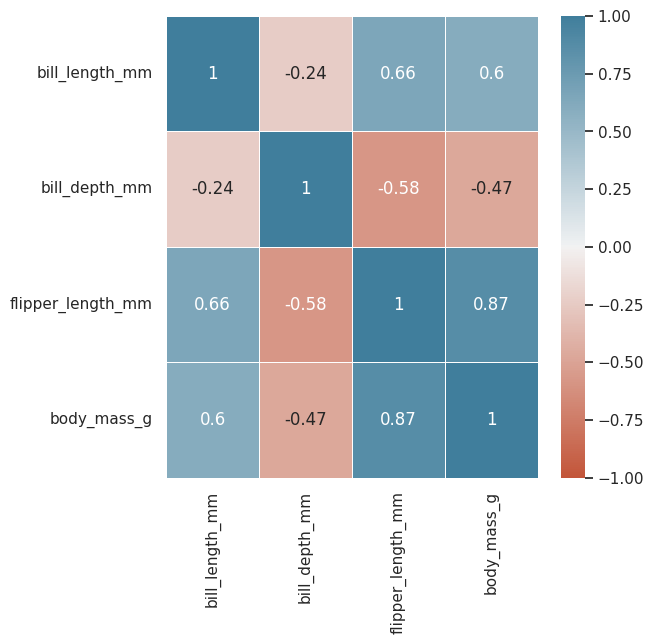
df.drop(["year"], axis=1).corr() #first remove the year column and then run
/tmp/ipykernel_1302/2116061803.py:1: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
df.drop(["year"], axis=1).corr() #first remove the year column and then run
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| bill_length_mm | 1.000000 | -0.235053 | 0.656181 | 0.595110 |
| bill_depth_mm | -0.235053 | 1.000000 | -0.583851 | -0.471916 |
| flipper_length_mm | 0.656181 | -0.583851 | 1.000000 | 0.871202 |
| body_mass_g | 0.595110 | -0.471916 | 0.871202 | 1.000000 |
Flipper length and body mass have a 0.8712 correlation coef, high
let’s visualize that into a matrix
plt.figure(figsize=(6, 6))
sns.heatmap(
data=df.drop(["year"], axis=1).corr(),
cmap=sns.diverging_palette(20, 230, as_cmap=True), #nice color palette
center=0, #centering the color legend
vmin=-1, #set min corr value to -1
vmax=1, #set max corr value to 1
linewidths=0.5,
annot=True
)
/tmp/ipykernel_1302/2892716928.py:3: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
data=df.drop(["year"], axis=1).corr(),
<AxesSubplot: >
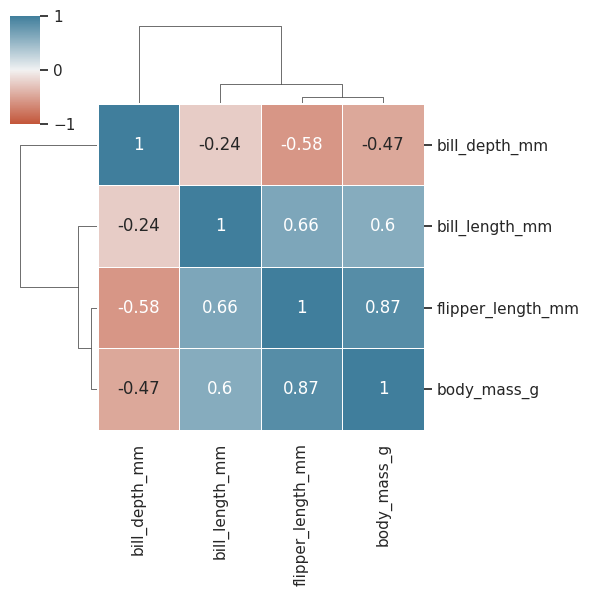
sns.clustermap(
data=df.drop(["year"], axis=1).corr(),
cmap=sns.diverging_palette(20, 230, as_cmap=True), #nice color palette
center=0, #centering the color legend
vmin=-1, #set min corr value to -1
vmax=1, #set max corr value to 1
linewidths=0.5,
annot=True,
figsize=(6,6)
);
/tmp/ipykernel_1302/2652653307.py:2: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
data=df.drop(["year"], axis=1).corr(),
How can i represent a categorical variable as a discrete numerical?#
Let’s do it with sex, set female to 0 and male to 1
df = df.assign(
numeric_sex=lambda df : df.sex.replace(["female", "male"], [0, 1])
)
df_clean = df.dropna()
sns.clustermap(
data=df.drop(["year"], axis=1).corr(),
cmap=sns.diverging_palette(20, 230, as_cmap=True), #nice color palette
center=0, #centering the color legend
vmin=-1, #set min corr value to -1
vmax=1, #set max corr value to 1
linewidths=0.5,
annot=True,
figsize=(6,6)
);
/tmp/ipykernel_1302/2652653307.py:2: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
data=df.drop(["year"], axis=1).corr(),
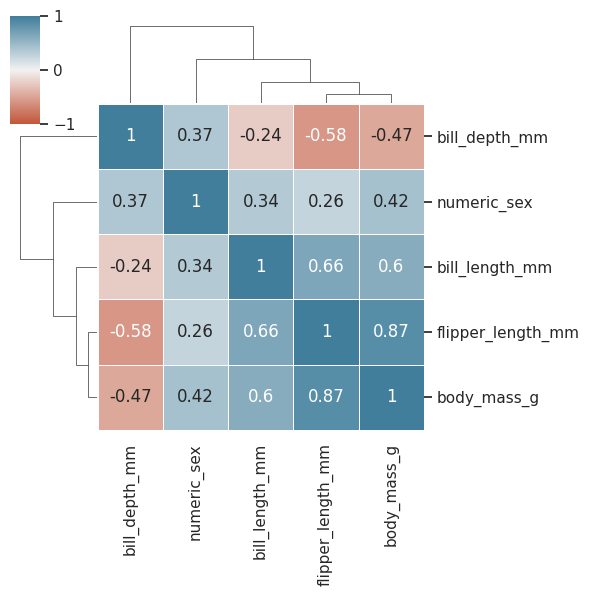
Look like linear correlation is low among sex and the other variables
Multiple regression#
I forgot the scale to weigh the penguins, what is the best way to capture that data?
Let’s create many linear regression models and compare them
Use the df_clean, which contains the dataset without null values, just to avoid errors
Model 1#
let’s create a model to predict the “body_mass” using the variable “bill_depth”
#Creating the model
model_1 = (
smf.ols(
formula="body_mass_g ~ bill_length_mm",
data=df_clean
)
.fit()
)
#Describing the model
model_1.summary()
| Dep. Variable: | body_mass_g | R-squared: | 0.347 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.345 |
| Method: | Least Squares | F-statistic: | 176.2 |
| Date: | Mon, 09 Jan 2023 | Prob (F-statistic): | 1.54e-32 |
| Time: | 21:34:02 | Log-Likelihood: | -2629.1 |
| No. Observations: | 333 | AIC: | 5262. |
| Df Residuals: | 331 | BIC: | 5270. |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 388.8452 | 289.817 | 1.342 | 0.181 | -181.271 | 958.961 |
| bill_length_mm | 86.7918 | 6.538 | 13.276 | 0.000 | 73.931 | 99.652 |
| Omnibus: | 6.141 | Durbin-Watson: | 0.849 |
|---|---|---|---|
| Prob(Omnibus): | 0.046 | Jarque-Bera (JB): | 4.899 |
| Skew: | -0.197 | Prob(JB): | 0.0864 |
| Kurtosis: | 2.555 | Cond. No. | 360. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Model 2#
Now let’s add a second variable “bill_depth_mm”
#Creating the model
model_2 = (
smf.ols(
formula="body_mass_g ~ bill_length_mm + bill_depth_mm",
data=df_clean
)
.fit()
)
#Describing the model
model_2.summary()
| Dep. Variable: | body_mass_g | R-squared: | 0.467 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.464 |
| Method: | Least Squares | F-statistic: | 144.8 |
| Date: | Mon, 09 Jan 2023 | Prob (F-statistic): | 7.04e-46 |
| Time: | 21:34:02 | Log-Likelihood: | -2595.2 |
| No. Observations: | 333 | AIC: | 5196. |
| Df Residuals: | 330 | BIC: | 5208. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 3413.4519 | 437.911 | 7.795 | 0.000 | 2552.002 | 4274.902 |
| bill_length_mm | 74.8126 | 6.076 | 12.313 | 0.000 | 62.860 | 86.765 |
| bill_depth_mm | -145.5072 | 16.873 | -8.624 | 0.000 | -178.699 | -112.315 |
| Omnibus: | 2.839 | Durbin-Watson: | 1.798 |
|---|---|---|---|
| Prob(Omnibus): | 0.242 | Jarque-Bera (JB): | 2.175 |
| Skew: | -0.000 | Prob(JB): | 0.337 |
| Kurtosis: | 2.604 | Cond. No. | 644. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Model 3#
Now we can add the “flipper_length_mm” and see what happens
#Creating the model
model_3 = (
smf.ols(
formula="body_mass_g ~ bill_length_mm + bill_depth_mm + flipper_length_mm",
data=df_clean
)
.fit()
)
#Describing the model
model_3.summary()
| Dep. Variable: | body_mass_g | R-squared: | 0.764 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.762 |
| Method: | Least Squares | F-statistic: | 354.9 |
| Date: | Mon, 09 Jan 2023 | Prob (F-statistic): | 9.26e-103 |
| Time: | 21:34:02 | Log-Likelihood: | -2459.8 |
| No. Observations: | 333 | AIC: | 4928. |
| Df Residuals: | 329 | BIC: | 4943. |
| Df Model: | 3 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -6445.4760 | 566.130 | -11.385 | 0.000 | -7559.167 | -5331.785 |
| bill_length_mm | 3.2929 | 5.366 | 0.614 | 0.540 | -7.263 | 13.849 |
| bill_depth_mm | 17.8364 | 13.826 | 1.290 | 0.198 | -9.362 | 45.035 |
| flipper_length_mm | 50.7621 | 2.497 | 20.327 | 0.000 | 45.850 | 55.675 |
| Omnibus: | 5.596 | Durbin-Watson: | 1.982 |
|---|---|---|---|
| Prob(Omnibus): | 0.061 | Jarque-Bera (JB): | 5.469 |
| Skew: | 0.312 | Prob(JB): | 0.0649 |
| Kurtosis: | 3.068 | Cond. No. | 5.44e+03 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 5.44e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
Model 4#
#Creating the model
model_4 = (
smf.ols(
formula="body_mass_g ~ bill_length_mm + bill_depth_mm + flipper_length_mm + C(sex)",
data=df_clean
)
.fit()
)
#Describing the model
model_4.summary()
| Dep. Variable: | body_mass_g | R-squared: | 0.823 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.821 |
| Method: | Least Squares | F-statistic: | 381.3 |
| Date: | Mon, 09 Jan 2023 | Prob (F-statistic): | 6.28e-122 |
| Time: | 21:34:02 | Log-Likelihood: | -2411.8 |
| No. Observations: | 333 | AIC: | 4834. |
| Df Residuals: | 328 | BIC: | 4853. |
| Df Model: | 4 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -2288.4650 | 631.580 | -3.623 | 0.000 | -3530.924 | -1046.006 |
| C(sex)[T.male] | 541.0285 | 51.710 | 10.463 | 0.000 | 439.304 | 642.753 |
| bill_length_mm | -2.3287 | 4.684 | -0.497 | 0.619 | -11.544 | 6.886 |
| bill_depth_mm | -86.0882 | 15.570 | -5.529 | 0.000 | -116.718 | -55.459 |
| flipper_length_mm | 38.8258 | 2.448 | 15.862 | 0.000 | 34.011 | 43.641 |
| Omnibus: | 2.598 | Durbin-Watson: | 1.843 |
|---|---|---|---|
| Prob(Omnibus): | 0.273 | Jarque-Bera (JB): | 2.125 |
| Skew: | 0.062 | Prob(JB): | 0.346 |
| Kurtosis: | 2.629 | Cond. No. | 7.01e+03 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 7.01e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
Model 5#
Let’s now try with the “flipper_length_mm” alone
#Creating the model
model_5 = (
smf.ols(
formula="body_mass_g ~ flipper_length_mm + C(sex)",
data=df_clean
)
.fit()
)
#Describing the model
model_5.summary()
| Dep. Variable: | body_mass_g | R-squared: | 0.806 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.805 |
| Method: | Least Squares | F-statistic: | 684.8 |
| Date: | Mon, 09 Jan 2023 | Prob (F-statistic): | 3.53e-118 |
| Time: | 21:34:02 | Log-Likelihood: | -2427.2 |
| No. Observations: | 333 | AIC: | 4860. |
| Df Residuals: | 330 | BIC: | 4872. |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -5410.3002 | 285.798 | -18.931 | 0.000 | -5972.515 | -4848.085 |
| C(sex)[T.male] | 347.8503 | 40.342 | 8.623 | 0.000 | 268.491 | 427.209 |
| flipper_length_mm | 46.9822 | 1.441 | 32.598 | 0.000 | 44.147 | 49.817 |
| Omnibus: | 0.262 | Durbin-Watson: | 1.710 |
|---|---|---|---|
| Prob(Omnibus): | 0.877 | Jarque-Bera (JB): | 0.376 |
| Skew: | 0.051 | Prob(JB): | 0.829 |
| Kurtosis: | 2.870 | Cond. No. | 2.95e+03 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 2.95e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
Note that only one variable explains the 76% of the variance.
You decide if sticking with one variable and getting 76% of variance, or stick with 4 and get 80%.
Multiple Regression Model Comparisons#
Creation of a results table#
model_results = pd.DataFrame(
dict(
actual_value = df_clean.body_mass_g,
prediction_model_1 = model_1.predict(),
prediction_model_2 = model_2.predict(),
prediction_model_3 = model_3.predict(),
prediction_model_4 = model_4.predict(),
prediction_model_5 = model_5.predict(),
species = df_clean.species,
sex = df_clean.sex
)
)
model_results
| actual_value | prediction_model_1 | prediction_model_2 | prediction_model_3 | prediction_model_4 | prediction_model_5 | species | sex | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3750.0 | 3782.402961 | 3617.641192 | 3204.761227 | 3579.136946 | 3441.323750 | Adelie | male |
| 1 | 3800.0 | 3817.119665 | 3836.725580 | 3436.701722 | 3343.220772 | 3328.384372 | Adelie | female |
| 2 | 3250.0 | 3886.553073 | 3809.271371 | 3906.897032 | 3639.137335 | 3751.223949 | Adelie | female |
| 4 | 3450.0 | 3574.102738 | 3350.786581 | 3816.705772 | 3457.954243 | 3657.259599 | Adelie | female |
| 5 | 3650.0 | 3799.761313 | 3356.140070 | 3696.168128 | 3764.536023 | 3864.163327 | Adelie | male |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 339 | 4000.0 | 5231.825347 | 4706.954140 | 4599.187485 | 4455.022405 | 4662.860306 | Chinstrap | male |
| 340 | 3400.0 | 4164.286703 | 4034.121055 | 4274.552753 | 3894.857519 | 4080.099176 | Chinstrap | female |
| 341 | 3775.0 | 4693.716437 | 4475.927353 | 3839.563668 | 4063.639819 | 4005.109853 | Chinstrap | male |
| 342 | 4100.0 | 4797.866549 | 4449.296758 | 4720.740455 | 4652.013882 | 4803.806832 | Chinstrap | male |
| 343 | 3775.0 | 4745.791493 | 4448.061337 | 4104.268240 | 3672.299099 | 3892.170475 | Chinstrap | female |
333 rows × 8 columns
Comparing predicted values vs Actual values (ECDFs)#
plt.figure(figsize=(6, 6))
sns.ecdfplot(
data=model_results
);
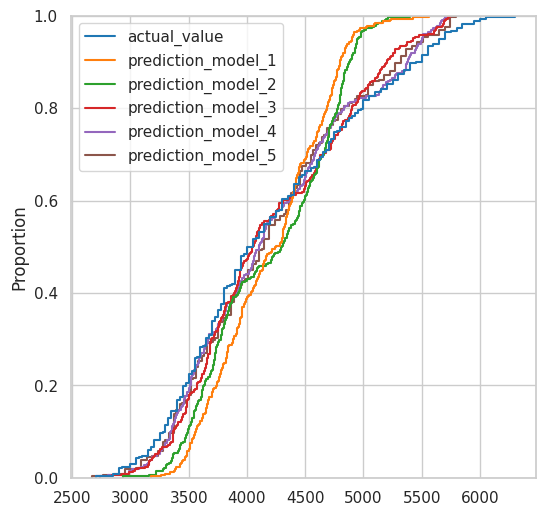
Now, note that the BLUE LINE ARE THE ACTUAL VALUES
Therefore, the line that is closer to the model, would be the better model
Comparing predicted values distributions vs Original values distributions(Kdeplot)#
plt.figure(figsize=(6, 6))
sns.kdeplot(
data=model_results
);
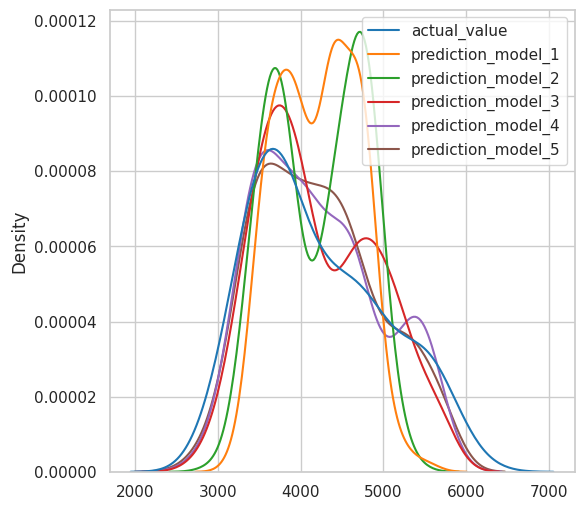
Looks like the purple line (model 4) fits better, but that contains 4 variables.
The model 5 also fist really great with only two variables
Bonus: Pandas Profiling Report#
import palmerpenguins
import pandas as pd
import pandas_profiling as pp
df = palmerpenguins.load_penguins()
pp.ProfileReport(df)
